Start at 2015-07-23 12:19:50
First, load the ontology 'HPPA' (2015-07-23 12:19:50) ...
'onto.HPPA' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment
Second, import files for annotations 'http://dcgor.r-forge.r-project.org/data/Algo/HP_anno.txt' and architectures 'http://dcgor.r-forge.r-project.org/data/Algo/SCOP_architecture.txt' (2015-07-23 12:19:50) ...
Third, propagate annotations (2015-07-23 12:19:51) ...
At level 16, there are 2 nodes, and 5 incoming neighbors.
At level 15, there are 7 nodes, and 9 incoming neighbors.
At level 14, there are 21 nodes, and 42 incoming neighbors.
At level 13, there are 54 nodes, and 82 incoming neighbors.
At level 12, there are 105 nodes, and 105 incoming neighbors.
At level 11, there are 274 nodes, and 188 incoming neighbors.
At level 10, there are 463 nodes, and 294 incoming neighbors.
At level 9, there are 782 nodes, and 441 incoming neighbors.
At level 8, there are 1004 nodes, and 538 incoming neighbors.
At level 7, there are 1182 nodes, and 581 incoming neighbors.
At level 6, there are 1295 nodes, and 527 incoming neighbors.
At level 5, there are 940 nodes, and 290 incoming neighbors.
At level 4, there are 408 nodes, and 99 incoming neighbors.
At level 3, there are 114 nodes, and 21 incoming neighbors.
At level 2, there are 21 nodes, and 1 incoming neighbors.
At level 1, there are 1 nodes, and 0 incoming neighbors.
There are 6673 terms used (2015-07-23 12:21:06).
Fourth, define groups using feature mode 'supra' (2015-07-23 12:21:06) ...
i) split into features (2015-07-23 12:21:06) ...
1 out of 4351 (2015-07-23 12:21:06)
436 out of 4351 (2015-07-23 12:21:06)
872 out of 4351 (2015-07-23 12:21:06)
1308 out of 4351 (2015-07-23 12:21:07)
1744 out of 4351 (2015-07-23 12:21:07)
2180 out of 4351 (2015-07-23 12:21:07)
2616 out of 4351 (2015-07-23 12:21:07)
3052 out of 4351 (2015-07-23 12:21:08)
3488 out of 4351 (2015-07-23 12:21:08)
3924 out of 4351 (2015-07-23 12:21:08)
4351 out of 4351 (2015-07-23 12:21:08)
ii) obtain feature-based groups (2015-07-23 12:21:08) ...
1 out of 4351 (2015-07-23 12:21:08)
436 out of 4351 (2015-07-23 12:21:08)
872 out of 4351 (2015-07-23 12:21:08)
1308 out of 4351 (2015-07-23 12:21:08)
1744 out of 4351 (2015-07-23 12:21:08)
2180 out of 4351 (2015-07-23 12:21:08)
2616 out of 4351 (2015-07-23 12:21:08)
3052 out of 4351 (2015-07-23 12:21:08)
3488 out of 4351 (2015-07-23 12:21:08)
3924 out of 4351 (2015-07-23 12:21:08)
4351 out of 4351 (2015-07-23 12:21:08)
There are 2194 features used (2015-07-23 12:21:09).
Finally, estimate associations between 2194 features and 6673 terms, with 3 min overlaps and 5.0e-02 fdr cutoff (2015-07-23 12:21:09) ...
1 out of 2194 (2015-07-23 12:21:12)
220 out of 2194 (2015-07-23 12:21:13)
440 out of 2194 (2015-07-23 12:21:14)
660 out of 2194 (2015-07-23 12:21:15)
880 out of 2194 (2015-07-23 12:21:16)
1100 out of 2194 (2015-07-23 12:21:17)
1320 out of 2194 (2015-07-23 12:21:18)
1540 out of 2194 (2015-07-23 12:21:19)
1760 out of 2194 (2015-07-23 12:21:19)
1980 out of 2194 (2015-07-23 12:21:20)
2194 out of 2194 (2015-07-23 12:21:21)
The results have been saved into '/Users/hfang/Sites/SUPERFAMILY/dcGO/dcGOR/sf2HPPA.txt'.
End at 2015-07-23 12:21:21
Runtime in total is: 91 secs
res[1:5,]
Feature_id Term_id Score
1 100895 HP:0004370 3.00
2 100895 HP:0002027 4.40
3 100895 HP:0011123 0.82
4 100895 HP:0004295 5.00
5 100895 HP:0002024 2.20
Start at 2015-07-23 12:21:21
Read the input file 'sf2HPPA.txt' (2015-07-23 12:21:21) ...
Load the ontology 'HPPA' (2015-07-23 12:21:21) ...
'onto.HPPA' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment
Do propagation via 'max' operation (2015-07-23 12:21:24) ...
At level 14, there are 1 nodes, and 3 incoming neighbors (2015-07-23 12:21:25).
At level 13, there are 2 nodes, and 7 incoming neighbors (2015-07-23 12:21:25).
At level 12, there are 10 nodes, and 18 incoming neighbors (2015-07-23 12:21:25).
At level 11, there are 19 nodes, and 26 incoming neighbors (2015-07-23 12:21:25).
At level 10, there are 68 nodes, and 74 incoming neighbors (2015-07-23 12:21:25).
At level 9, there are 118 nodes, and 100 incoming neighbors (2015-07-23 12:21:25).
At level 8, there are 174 nodes, and 137 incoming neighbors (2015-07-23 12:21:26).
At level 7, there are 240 nodes, and 181 incoming neighbors (2015-07-23 12:21:27).
At level 6, there are 290 nodes, and 180 incoming neighbors (2015-07-23 12:21:27).
At level 5, there are 300 nodes, and 134 incoming neighbors (2015-07-23 12:21:28).
At level 4, there are 172 nodes, and 65 incoming neighbors (2015-07-23 12:21:29).
At level 3, there are 76 nodes, and 21 incoming neighbors (2015-07-23 12:21:29).
At level 2, there are 21 nodes, and 1 incoming neighbors (2015-07-23 12:21:29).
after propagation, there are 460 features annotated by 1492 terms.
Determining IC-based slim levels (2015-07-23 12:21:29) ...
1 level with 16 terms with IC falling around 0.33 (between 0.00 and 0.67).
2 level with 62 terms with IC falling around 1.00 (between 0.83 and 1.16).
3 level with 149 terms with IC falling around 1.66 (between 1.50 and 1.83).
4 level with 267 terms with IC falling around 2.33 (between 2.16 and 2.50).
An object of S3 class 'HIS' has been built and saved into '/Users/hfang/Sites/SUPERFAMILY/dcGO/dcGOR/sf2HPPA.HIS.RData'.
End at 2015-07-23 12:23:18
Runtime in total is: 117 secs
[1] "hscore" "ic" "slim"
Start at 2015-07-23 12:23:18
Read the input file 'http://dcgor.r-forge.r-project.org/data/Algo/SCOP_architecture.txt' ...
Predictions for 17467 sequences (with 4351 distinct architectures) using 'sf2HPPA.HIS.RData' RData, 'sum' merge method, 'log' scale method and 'supra' feature mode (2015-07-23 12:23:18) ...
##############################
'dcAlgoPredict' is being called...
##############################
Start at 2015-07-23 12:23:18
Load the customised HIS object 'sf2HPPA.HIS.RData' (2015-07-23 12:23:18)...
Predictions for 4351 architectures using 'sum' merge method, 'log' scale method and 'supra' feature mode (2015-07-23 12:23:18)...
1 out of 4351 (2015-07-23 12:23:18)
436 out of 4351 (2015-07-23 12:23:30)
872 out of 4351 (2015-07-23 12:23:31)
1308 out of 4351 (2015-07-23 12:23:32)
1744 out of 4351 (2015-07-23 12:23:33)
2180 out of 4351 (2015-07-23 12:23:34)
2616 out of 4351 (2015-07-23 12:23:35)
3052 out of 4351 (2015-07-23 12:23:36)
3488 out of 4351 (2015-07-23 12:23:37)
3924 out of 4351 (2015-07-23 12:23:37)
4351 out of 4351 (2015-07-23 12:23:39)
End at 2015-07-23 12:23:39
Runtime in total is: 21 secs
##############################
'dcAlgoPredict' has been completed!
##############################
Preparations for output (2015-07-23 12:23:39)...
The predictions have been saved into '/Users/hfang/Sites/SUPERFAMILY/dcGO/dcGOR/SCOP_architecture.HPPA_predicted.txt'.
End at 2015-07-23 12:23:40
Runtime in total is: 22 secs
res_Pred[1:5,]
SeqID Term Score
[1,] "100288966" "HP:0000118" "1"
[2,] "100288966" "HP:0000924" "1"
[3,] "100288966" "HP:0001760" "1"
[4,] "100288966" "HP:0001780" "1"
[5,] "100288966" "HP:0002813" "1"
Start at 2015-07-23 12:23:40
First, load the ontology 'HPPA' (2015-07-23 12:23:40) ...
'onto.HPPA' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment
Second, import files for GSP and predictions (2015-07-23 12:23:40) ...
Third, propagate GSP annotations (2015-07-23 12:23:44) ...
At level 16, there are 2 nodes, and 5 incoming neighbors.
At level 15, there are 7 nodes, and 9 incoming neighbors.
At level 14, there are 21 nodes, and 42 incoming neighbors.
At level 13, there are 54 nodes, and 82 incoming neighbors.
At level 12, there are 105 nodes, and 105 incoming neighbors.
At level 11, there are 274 nodes, and 188 incoming neighbors.
At level 10, there are 463 nodes, and 294 incoming neighbors.
At level 9, there are 782 nodes, and 441 incoming neighbors.
At level 8, there are 1004 nodes, and 538 incoming neighbors.
At level 7, there are 1182 nodes, and 581 incoming neighbors.
At level 6, there are 1295 nodes, and 527 incoming neighbors.
At level 5, there are 940 nodes, and 290 incoming neighbors.
At level 4, there are 408 nodes, and 99 incoming neighbors.
At level 3, there are 114 nodes, and 21 incoming neighbors.
At level 2, there are 21 nodes, and 1 incoming neighbors.
At level 1, there are 1 nodes, and 0 incoming neighbors.
There are 3048 genes/proteins in GSP (2015-07-23 12:24:32).
Fourth, process input predictions (2015-07-23 12:24:32) ...
There are 7426 genes/proteins in predictions (2015-07-23 12:24:33).
Fifth, calculate the precision and recall for each of 1694 predicted and GSP genes/proteins (2015-07-23 12:24:33).
Finally, calculate the averaged precision and recall (2015-07-23 12:24:34).
In summary, Prediction coverage: 0.56 (amongst 3048 targets in GSP), and F-measure: 0.26.
End at 2015-07-23 12:24:34
Runtime in total is: 54 secs
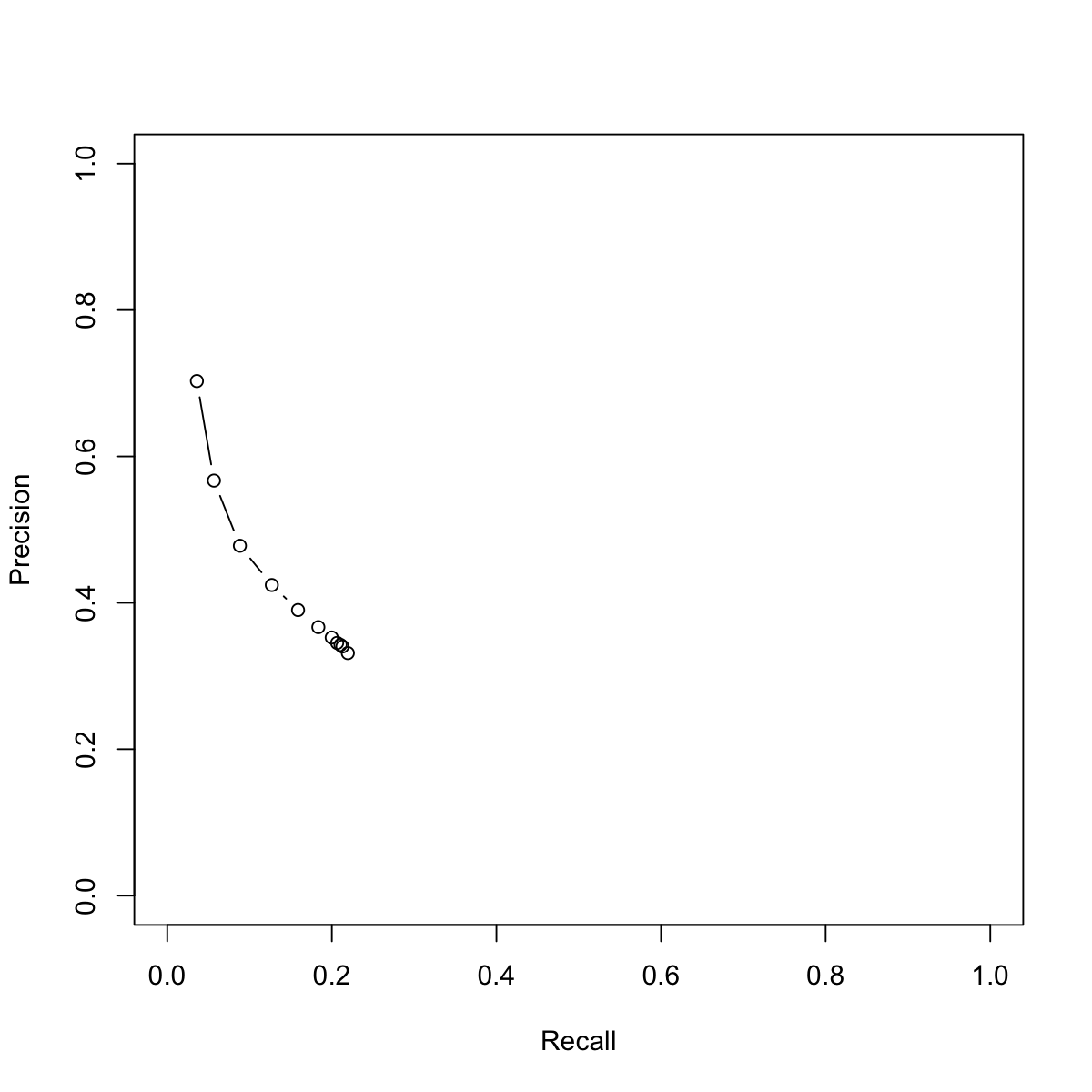
res_PR
Precision Recall
1 0.7029713 0.03603475
0.90001 0.5669893 0.05676330
0.80002 0.4779550 0.08826593
0.70003 0.4242904 0.12708065
0.60004 0.3901488 0.15893078
0.50005 0.3667623 0.18361571
0.40006 0.3526200 0.19984090
0.30007 0.3452423 0.20644254
0.20008 0.3425576 0.21048189
0.10009 0.3403761 0.21287068
1e-04 0.3313322 0.21942770
### plot PR curve
plot(res_PR[,2], res_PR[,1], xlim=c(0,1), ylim=c(0,1), type="b", xlab="Recall", ylab="Precision")
 #---------------------------------------------------------------------------
#---------------------------------------------------------------------------
# Compared to RWR-based predictions and naive predictions
#---------------------------------------------------------------------------
#---------------------------------------------------------------------------
## 1) RWR-based predictions, please see: dcRWRpredict
## 2) Naive predictions, please see: dcNaivePredict
#---------------------------------------------------------------------------
#---------------------------------------------------------------------------
# Compared to RWR-based predictions and naive predictions
#---------------------------------------------------------------------------
#---------------------------------------------------------------------------
## 1) RWR-based predictions, please see: dcRWRpredict
## 2) Naive predictions, please see: dcNaivePredict