Function to perform RWR-based ontology term predictions from input known annotations and an input graph
Description
dcRWRpredict is supposed to perform ontology term predictions
based on Random Walk with Restart (RWR) from input known annotations
and an input graph.
Usage
dcRWRpredict(data, g, output.file = NULL, ontology = c(NA, "GOBP", "GOMF", "GOCC",
"DO", "HPPA", "HPMI", "HPON", "MP", "EC", "KW", "UP"), method = c("indirect",
"direct"), normalise = c("laplacian", "row", "column", "none"), restart = 0.75,
normalise.affinity.matrix = c("none", "quantile"), leave.one.out = T, propagation = c("max",
"sum"), scale.method = c("log", "linear", "none"), parallel = TRUE, multicores = NULL,
verbose = T, RData.ontology.customised = NULL, RData.location = "https://github.com/hfang-bristol/RDataCentre/blob/master/dcGOR")
Arguments
- data
- an input gene-term data matrix containing known annotations used for seeds. Each value in input matrix does not necessarily have to be binary (non-zeros will be used as a weight, but should be non-negative for easy interpretation). Also, data can be a list, each containing the known annotated genes
- g
- an object of class "igraph" or
Dnetwork - output.file
- an output file containing predicted results. If not NULL, a tab-delimited text file will be also written out; otherwise, there is no output file (by default)
- ontology
- the ontology identity. It can be "GOBP" for Gene
Ontology Biological Process, "GOMF" for Gene Ontology Molecular
Function, "GOCC" for Gene Ontology Cellular Component, "DO" for Disease
Ontology, "HPPA" for Human Phenotype Phenotypic Abnormality, "HPMI" for
Human Phenotype Mode of Inheritance, "HPON" for Human Phenotype ONset
and clinical course, "MP" for Mammalian Phenotype, "EC" for Enzyme
Commission, "KW" for UniProtKB KeyWords, "UP" for UniProtKB UniPathway.
For details on the eligibility for pairs of input domain and ontology,
please refer to the online Documentations at
http://supfam.org/dcGOR/docs.html. If NA, then the user has to
input a customised RData-formatted file (see
RData.ontology.customisedbelow) - method
- the method used to calculate RWR. It can be 'direct' for directly applying RWR, 'indirect' for indirectly applying RWR (first pre-compute affinity matrix and then derive the affinity score)
- normalise
- the way to normalise the adjacency matrix of the input graph. It can be 'laplacian' for laplacian normalisation, 'row' for row-wise normalisation, 'column' for column-wise normalisation, or 'none'
- restart
- the restart probability used for RWR. The restart probability takes the value from 0 to 1, controlling the range from the starting nodes/seeds that the walker will explore. The higher the value, the more likely the walker is to visit the nodes centered on the starting nodes. At the extreme when the restart probability is zero, the walker moves freely to the neighbors at each step without restarting from seeds, i.e., following a random walk (RW)
- normalise.affinity.matrix
- the way to normalise the output affinity matrix. It can be 'none' for no normalisation, 'quantile' for quantile normalisation to ensure that columns (if multiple) of the output affinity matrix have the same quantiles
- leave.one.out
- logical to indicate whether the leave-one-out test is used for predictions. By default, it sets to true for doing leave-one-out test (that is, known seeds are removed)
- propagation
- how to propagate the score. It can be "max" for retaining the maximum score (by default), "sum" for additively accumulating the score
- scale.method
- the method used to scale the predictive scores. It
can be: "none" for no scaling, "linear" for being linearily scaled into
the range between 0 and 1, "log" for the same as "linear" but being
first log-transformed before being scaled. The scaling between 0 and 1
is done via:
\frac{S - S_{min}}{S_{max} - S_{min}}, whereS_{min}andS_{max}are the minimum and maximum values forS - parallel
- logical to indicate whether parallel computation with
multicores is used. By default, it sets to true, but not necessarily
does so. Partly because parallel backends available will be
system-specific (now only Linux or Mac OS). Also, it will depend on
whether these two packages "foreach" and "doMC" have been installed. It
can be installed via:
source("http://bioconductor.org/biocLite.R"); biocLite(c("foreach","doMC")). If not yet installed, this option will be disabled - multicores
- an integer to specify how many cores will be registered as the multicore parallel backend to the 'foreach' package. If NULL, it will use a half of cores available in a user's computer. This option only works when parallel computation is enabled
- verbose
- logical to indicate whether the messages will be displayed in the screen. By default, it sets to true for display
- RData.ontology.customised
- a file name for RData-formatted file
containing an object of S4 class 'Onto' (i.g. ontology). By default, it
is NULL. It is only needed when the user wants to perform customised
analysis using their own ontology. See
dcBuildOntofor how to creat this object - RData.location
- the characters to tell the location of built-in
RData files. By default, it remotely locates at
"https://github.com/hfang-bristol/RDataCentre/blob/master/dcGOR" and
"http://dcgor.r-forge.r-project.org/data". For the user equipped with
fast internet connection, this option can be just left as default. But
it is always advisable to download these files locally. Especially when
the user needs to run this function many times, there is no need to ask
the function to remotely download every time (also it will
unnecessarily increase the runtime). For examples, these files (as a
whole or part of them) can be first downloaded into your current
working directory, and then set this option as:
RData.location=".". If RData to load is already part of package itself, this parameter can be ignored (since this function will try to load it via functiondatafirst). Here is the UNIX command for downloading all RData files (preserving the directory structure):wget -r -l2 -A "*.RData" -np -nH --cut-dirs=0 "http://dcgor.r-forge.r-project.org/data"
Value
a data frame containing three columns: 1st column the same as the input file (e.g. 'SeqID'), 2nd for 'Term' (predicted ontology terms), 3rd for 'Score' (along with predicted scores)
Note
When 'output.file' is specified, a tab-delimited text file is written out, with the column names: 1st column the same as the input file (e.g. 'SeqID'), 2nd for 'Term' (predicted ontology terms), 3rd for 'Score' (along with predicted scores). The choice of which method to use RWR depends on the number of seed sets and whether using leave-one-out test. If the total product of both numbers are huge, it is better to use 'indrect' method (for a single run). Also, when using leave-one-out test, it has to be use 'indrect' method.
Examples
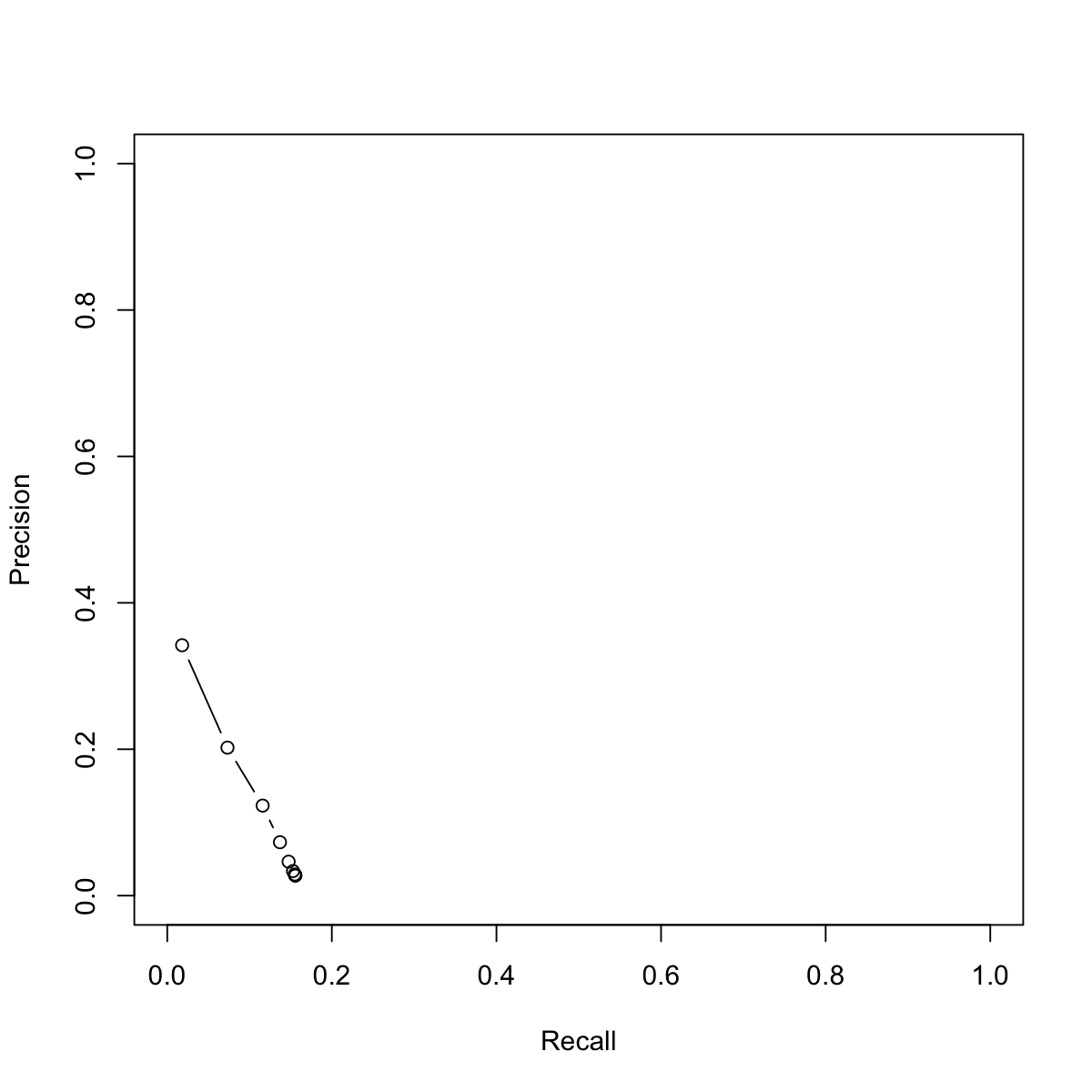
# 1) define an input network ## 1a) an igraph object that contains a functional protein association network in human. ### The network is extracted from the STRING database (version 9.1). ### Only those associations with medium confidence (score>=400) are retained org.Hs.string <- dnet::dRDataLoader(RData='org.Hs.string')'org.Hs.string' (from https://github.com/hfang-bristol/RDataCentre/blob/master/dnet/1.0.7/org.Hs.string.RData?raw=true) has been loaded into the working environment (at 2015-07-23 12:59:02)## 1b) restrict to those edges with confidence score>=999 ### keep the largest connected component network <- igraph::subgraph.edges(org.Hs.string, eids=E(org.Hs.string)[combined_score>=999]) g <- dnet::dNetInduce(g=network, nodes_query=V(network)$name, largest.comp=TRUE) ## Notably, in reality, 1b) can be replaced by: #g <- igraph::subgraph.edges(org.Hs.string, eids=E(org.Hs.string)[combined_score>=400]) ## 1c) make sure there is a 'weight' edge attribute E(g)$weight <- E(g)$combined_score ### use EntrezGene ID as default 'name' node attribute V(g)$name <- V(g)$geneid gIGRAPH UNW- 2071 7603 -- + attr: name (v/c), seqid (v/c), geneid (v/n), symbol (v/c), | description (v/c), neighborhood_score (e/n), fusion_score (e/n), | cooccurence_score (e/n), coexpression_score (e/n), experimental_score | (e/n), database_score (e/n), textmining_score (e/n), combined_score | (e/n), weight (e/n) + edges (vertex names): [1] 2288 --3320 1080 --3312 1080 --9368 1080 --7311 1407 --6500 [6] 1407 --8864 1407 --5187 1407 --8454 1407 --26224 1407 --1454 [11] 1407 --8863 1407 --8945 1407 --406 5536 --3320 2067 --4436 [16] 2072 --2067 2067 --7507 9821 --8408 9776 --9821 10533--11345 + ... omitted several edges# 2) define the known annotations as seeds anno.file <- "http://dcgor.r-forge.r-project.org/data/Algo/HP_anno.txt" data <- dcSparseMatrix(anno.file)Read the input file 'http://dcgor.r-forge.r-project.org/data/Algo/HP_anno.txt' ... There are 80781 entries, converted into a sparse matrix of 3085 X 5715.# 3) perform RWR-based ontology term predictions res <- dcRWRpredict(data=data, g=g, ontology="HPPA", parallel=FALSE)Start at 2015-07-23 12:59:08 First, RWR on input graph (2071 nodes and 7603 edges) using input matrix (3085 rows and 5715 columns) as seeds (2015-07-23 12:59:08)... using 'indirect' method to do RWR with leave-one-out test (2015-07-23 12:59:08)... ############################## 'dnet::dRWR' is being called... ############################## Start at 2015-07-23 12:59:08 First, get the adjacency matrix of the input graph (2015-07-23 12:59:08) ... Notes: using weighted graph! Then, normalise the adjacency matrix using laplacian normalisation (2015-07-23 12:59:08) ... Third, RWR of 2071 sets of seeds using 7.5e-01 restart probability (2015-07-23 12:59:09) ... 1 out of 2071 seed sets (2015-07-23 12:59:09) 208 out of 2071 seed sets (2015-07-23 12:59:23) 416 out of 2071 seed sets (2015-07-23 12:59:38) 624 out of 2071 seed sets (2015-07-23 12:59:54) 832 out of 2071 seed sets (2015-07-23 13:00:11) 1040 out of 2071 seed sets (2015-07-23 13:00:31) 1248 out of 2071 seed sets (2015-07-23 13:00:52) 1456 out of 2071 seed sets (2015-07-23 13:01:16) 1664 out of 2071 seed sets (2015-07-23 13:01:42) 1872 out of 2071 seed sets (2015-07-23 13:02:11) 2071 out of 2071 seed sets (2015-07-23 13:02:39) Fourth, rescale steady probability vector (2015-07-23 13:02:39) ... Finally, output 2071 by 2071 affinity matrix normalised by none (2015-07-23 13:02:39) ... Finish at 2015-07-23 13:02:40 Runtime in total is: 212 secs ############################## 'dnet::dRWR' has been completed! ############################## Second, propagate 'HPPA' ontology annotations via 'max' operation (2015-07-23 13:02:44)...Second, propagate 'HPPA' ontology annotations via 'sum' operation (2015-07-23 13:02:44)... ############################## 'dcAlgoPropagate' is being called... ############################## Start at 2015-07-23 13:02:51 Load the input file (2015-07-23 13:02:51) ... Load the ontology 'HPPA' (2015-07-23 13:02:55) ... 'onto.HPPA' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment Do propagation via 'max' operation (2015-07-23 13:02:57) ... At level 16, there are 2 nodes, and 5 incoming neighbors (2015-07-23 13:02:59). At level 15, there are 5 nodes, and 9 incoming neighbors (2015-07-23 13:03:00). At level 14, there are 15 nodes, and 32 incoming neighbors (2015-07-23 13:03:00). At level 13, there are 36 nodes, and 60 incoming neighbors (2015-07-23 13:03:01). At level 12, there are 63 nodes, and 66 incoming neighbors (2015-07-23 13:03:03). At level 11, there are 125 nodes, and 114 incoming neighbors (2015-07-23 13:03:06). At level 10, there are 230 nodes, and 182 incoming neighbors (2015-07-23 13:03:11). At level 9, there are 396 nodes, and 279 incoming neighbors (2015-07-23 13:03:19). At level 8, there are 557 nodes, and 370 incoming neighbors (2015-07-23 13:03:29). At level 7, there are 667 nodes, and 381 incoming neighbors (2015-07-23 13:03:42). At level 6, there are 731 nodes, and 361 incoming neighbors (2015-07-23 13:03:56). At level 5, there are 583 nodes, and 220 incoming neighbors (2015-07-23 13:04:06). At level 4, there are 290 nodes, and 89 incoming neighbors (2015-07-23 13:04:11). At level 3, there are 100 nodes, and 21 incoming neighbors (2015-07-23 13:04:13). At level 2, there are 21 nodes, and 1 incoming neighbors (2015-07-23 13:04:13). after propagation, there are 2056 features annotated by 3822 terms. Determining IC-based slim levels (2015-07-23 13:04:24) ... 1 level with 647 terms with IC falling around 0.01 (between 0.00 and 0.02). 2 level with 0 terms with IC falling around 0.03 (between 0.03 and 0.04). 3 level with 0 terms with IC falling around 0.05 (between 0.05 and 0.06). 4 level with 0 terms with IC falling around 0.08 (between 0.07 and 0.08). End at 2015-07-23 13:15:13 Runtime in total is: 742 secs ############################## 'dcAlgoPropagate' has been completed! ############################## Third, rescale predictive score using 'log' method (2015-07-23 13:15:13)... The input list has been converted into a matrix of 7868533 X 3. Finish at 2015-07-23 13:15:23 Runtime in total is: 975 secsres[1:10,]SeqID Term Score [1,] "1000" "HP:0000118" "1" [2,] "1000" "HP:0000119" "0.9202" [3,] "1000" "HP:0000152" "0.9519" [4,] "1000" "HP:0000478" "0.9079" [5,] "1000" "HP:0000598" "0.9519" [6,] "1000" "HP:0000707" "0.8922" [7,] "1000" "HP:0000769" "0.8509" [8,] "1000" "HP:0000818" "0.8692" [9,] "1000" "HP:0000924" "0.9585" [10,] "1000" "HP:0001197" "0.8663"# 4) calculate Precision and Recall GSP.file <- anno.file prediction.file <- res res_PR <- dcAlgoPredictPR(GSP.file=GSP.file, prediction.file=prediction.file, ontology="HPPA")Start at 2015-07-23 13:15:23 First, load the ontology 'HPPA' (2015-07-23 13:15:23) ... 'onto.HPPA' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment Second, import files for GSP and predictions (2015-07-23 13:15:23) ... Third, propagate GSP annotations (2015-07-23 13:15:27) ... At level 16, there are 2 nodes, and 5 incoming neighbors. At level 15, there are 7 nodes, and 9 incoming neighbors. At level 14, there are 21 nodes, and 42 incoming neighbors. At level 13, there are 54 nodes, and 82 incoming neighbors. At level 12, there are 105 nodes, and 105 incoming neighbors. At level 11, there are 274 nodes, and 188 incoming neighbors. At level 10, there are 463 nodes, and 294 incoming neighbors. At level 9, there are 782 nodes, and 441 incoming neighbors. At level 8, there are 1004 nodes, and 538 incoming neighbors. At level 7, there are 1182 nodes, and 581 incoming neighbors. At level 6, there are 1295 nodes, and 527 incoming neighbors. At level 5, there are 940 nodes, and 290 incoming neighbors. At level 4, there are 408 nodes, and 99 incoming neighbors. At level 3, there are 114 nodes, and 21 incoming neighbors. At level 2, there are 21 nodes, and 1 incoming neighbors. At level 1, there are 1 nodes, and 0 incoming neighbors. There are 3048 genes/proteins in GSP (2015-07-23 13:16:12). Fourth, process input predictions (2015-07-23 13:16:12) ... There are 2056 genes/proteins in predictions (2015-07-23 13:16:24). Fifth, calculate the precision and recall for each of 488 predicted and GSP genes/proteins (2015-07-23 13:16:24). Finally, calculate the averaged precision and recall (2015-07-23 13:16:26). In summary, Prediction coverage: 0.16 (amongst 3048 targets in GSP), and F-measure: 0.12. End at 2015-07-23 13:16:26 Runtime in total is: 63 secsres_PRPrecision Recall 1 0.34207485 0.01806081 0.9 0.20221296 0.07321846 0.8 0.12302258 0.11590770 0.7 0.07294678 0.13697636 0.6 0.04637731 0.14749228 0.5 0.03358901 0.15280354 0.4 0.02898669 0.15491591 0.3 0.02810094 0.15542853 0.2 0.02776679 0.15556983 0.1 0.02763143 0.15561603 0 0.02758970 0.15562458# 5) Plot PR-curve plot(res_PR[,2], res_PR[,1], xlim=c(0,1), ylim=c(0,1), type="b", xlab="Recall", ylab="Precision")
Source code
dcRWRpredict.r
Source man
dcRWRpredict.Rd
dcRWRpredict.pdf
See also
dcRDataLoader, dcAlgoPropagate,
dcList2Matrix