Function to generate a subgraph of a direct acyclic graph (DAG) induced by the input annotation data
Description
dcDAGannotate is supposed to produce a subgraph induced by the
input annotation data, given a direct acyclic graph (DAG; an ontology).
The input is a graph of "igraph" or "Onto" object, a list of the
vertices containing annotation data, and the mode defining the paths to
the root of DAG. The induced subgraph contains vertices (with
annotation data) and their ancestors along with the defined paths to
the root of DAG. The annotations at these vertices (including their
ancestors) are also updated according to the true-path rule: a domain
annotated to a term should also be annotated by its all ancestor terms.
Usage
dcDAGannotate(g, annotations, path.mode = c("all_paths", "shortest_paths", "all_shortest_paths"),
verbose = TRUE)
Arguments
- g
- an object of class "igraph" or
Onto - annotations
- an object of class
Anno, that is, the vertices/nodes for which annotation data are provided - path.mode
- the mode of paths induced by vertices/nodes with input annotation data. It can be "all_paths" for all possible paths to the root, "shortest_paths" for only one path to the root (for each node in query), "all_shortest_paths" for all shortest paths to the root (i.e. for each node, find all shortest paths with the equal lengths)
- verbose
- logical to indicate whether the messages will be displayed in the screen. By default, it sets to true for display
Value
subg: an induced subgraph, an object of class "igraph" or "Onto" (the same as input). In addition to the original attributes to nodes and edges, the return subgraph is also appended by new node attributes: "annotations", which contains a list of domains either as original annotations or inherited annotations; "IC", which stands for information content defined as negative 10-based log-transformed frequency of domains annotated to that term.
Note
For the mode "shortest_paths", the induced subgraph is the most concise, and thus informative for visualisation when there are many nodes in query, while the mode "all_paths" results in the complete subgraph.
Examples
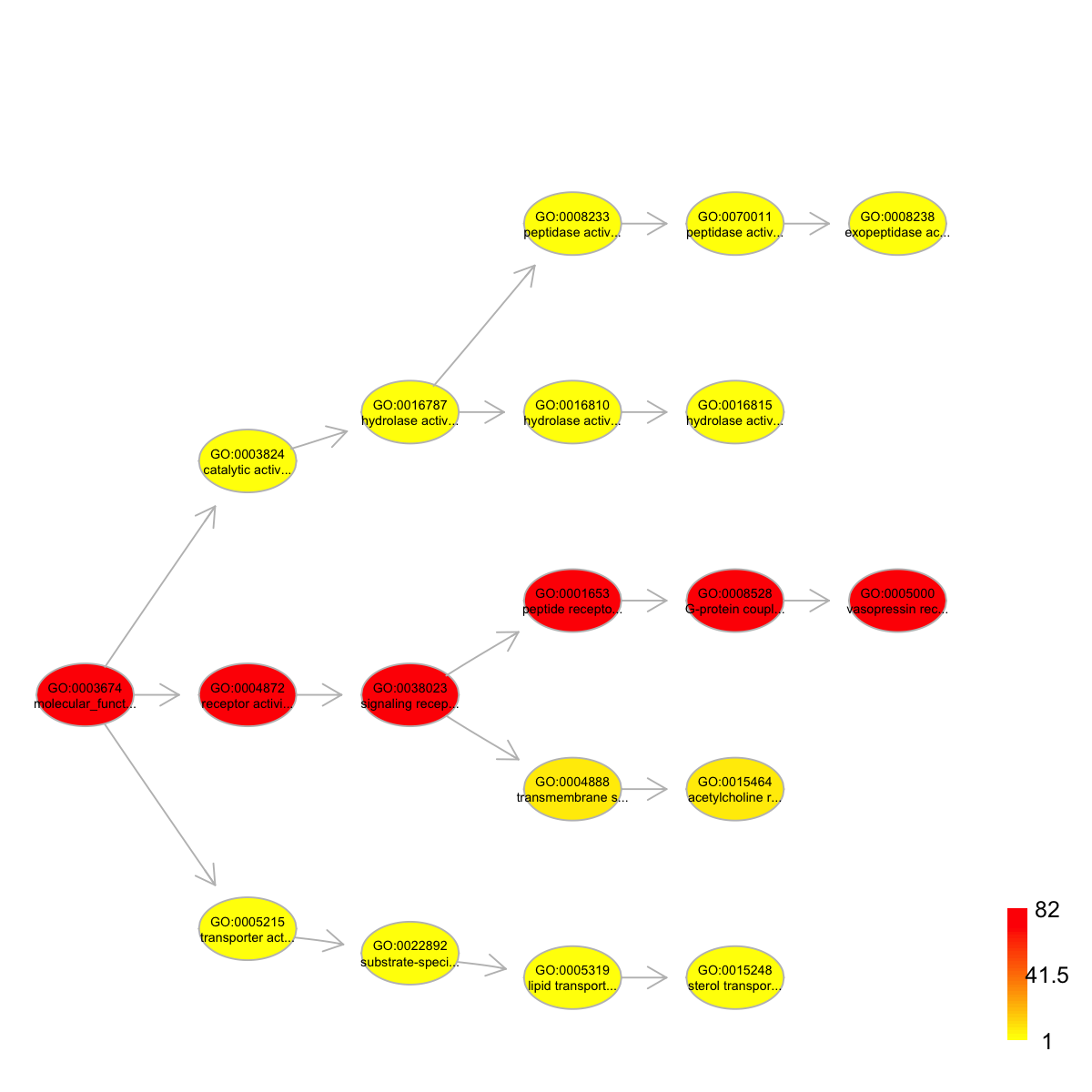
# 1) load onto.GOMF (as 'Onto' object) g <- dcRDataLoader('onto.GOMF')'onto.GOMF' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment# 2) load SCOP superfamilies annotated by GOMF (as 'Anno' object) Anno <- dcRDataLoader('SCOP.sf2GOMF')'SCOP.sf2GOMF' (from package 'dcGOR' version 1.0.5) has been loaded into the working environment# 3) prepare for annotation data # randomly select 5 terms vertices (and their annotation data) annotations <- Anno[,sample(1:dim(Anno)[2], 5)] # 4) obtain the induced subgraph according to the input annotation data # 4a) based on all possible paths (i.e. the complete subgraph induced) dcDAGannotate(g, annotations, path.mode="all_paths", verbose=TRUE)At level 8, there are 1 nodes, and 1 incoming neighbors. At level 7, there are 1 nodes, and 2 incoming neighbors. At level 6, there are 3 nodes, and 3 incoming neighbors. At level 5, there are 6 nodes, and 4 incoming neighbors. At level 4, there are 4 nodes, and 4 incoming neighbors. At level 3, there are 3 nodes, and 3 incoming neighbors. At level 2, there are 4 nodes, and 1 incoming neighbors. At level 1, there are 1 nodes, and 0 incoming neighbors.An object of S4 class 'Onto' @adjMatrix: a direct matrix of 23 terms (parents/from) X 23 terms (children/to) @nodeInfo (InfoDataFrame) nodeNames: GO:0003674 GO:0003824 GO:0004872 ... GO:0015464 GO:0005000 (23 total) nodeAttr: term_id term_name term_namespace term_distance annotations IC# 4b) based on shortest paths (i.e. the most concise subgraph induced) dag <- dcDAGannotate(g, annotations, path.mode="shortest_paths", verbose=TRUE)At level 6, there are 2 nodes, and 2 incoming neighbors. At level 5, there are 5 nodes, and 5 incoming neighbors. At level 4, there are 5 nodes, and 3 incoming neighbors. At level 3, there are 3 nodes, and 3 incoming neighbors. At level 2, there are 3 nodes, and 1 incoming neighbors. At level 1, there are 1 nodes, and 0 incoming neighbors.# 5) color-code nodes/terms according to the number of annotations if(class(dag)=='Onto') dag <- dcConverter(dag, from='Onto', to='igraph')Your input object 'dag' of class 'Onto' has been converted into an object of class 'igraph'.data <- sapply(V(dag)$annotations, length) names(data) <- V(dag)$name dnet::visDAG(g=dag, data=data, node.info="both")
Source code
dcDAGannotate.r
Source man
dcDAGannotate.Rd
dcDAGannotate.pdf
See also
dcRDataLoader, dcEnrichment,
dcDAGdomainSim, dcConverter